In this article, I will talk about my experience on scheduling data science project's notebooks on AWS Sagemaker instances using Airflow. We have been using Netflix's papermill library to run Jupyter notebooks more than 2 years now in production and everyday 10s of Sagemaker Notebook instances are orchestrated by Airflow working like a charm.
You will read about the general architectural design of this system, what is the way of working, what are the roles and responsibilities between teams and how you can implement it yourself.
The Problem
It all started with me reading this article on Netflix blog about running jupyter notebook files with external parameters for productionizing data science workloads. This could be the solution to a common problem which I faced in my previous company, we were running Apache Spark applications using pyspark and other python code for data science and reporting projects on AWS EMR. Most of the development was happening via notebooks since Jupyter notebooks are almost a standard development environment for data scientists. At that time, one common problem was to move code from ipynb notebook files to normal python files and back during development and preparing production code. Since those are 2 separate files, this involved a lot of copy pasting from notebooks to .py files for productionizing the work. While in the early stages of a project this was an easy task, later when an older project had to be modified for either new feature or bug fixing, you had to create a notebook file from python file every time or find the older notebook in your workspace and modify it with changes happened on python file. And when you are done with development and now it is time to put changes into python file, again copy paste recent changes you've made into the python file and test it. During this back and forth copy paste process or automatic generation of files, many times simple mistakes happened such as forgotten imports, one line changes missed, indentation errors, etc.. Some mistakes were immediately visible, some were only visible after running the spark app more than 3 hours. And this is the case only one person working on one project, imagine multiple data scientists are working in the same project with multiple notebook files, moving functions or code blocks from one file to another etc, it is a very complex job just to solve conflicts on git.
Another disadvantage of using notebooks for development, python for production is that you get a lot of "it was working on my development environment" response for errors that happened during testing of airflow automation dag. And these errors are simple coding mistakes, like I mentioned above, missing imports, hard-coded values, extra imports used during development, missing library in requirement.txt file and so on. So in essence maintainability of the projects was a pain point.
The Solution
When I started working at Leaseplan, I did not want to have the same issues and from the beginning looked for a way to establish a solid way of working between Data Science and Data Engineering teams. The idea was that data scientists are free to develop their project on Sagemaker notebook instances as they want as long as they deliver this project in a git repository with notebook files to run and whole dependencies in a requirements.txt file when it is time to productionize project using Airflow.
So from the data engineering perspective we only need to know which dependencies should be installed and then which notebook files have to be executed with a set of parameters and the order of notebook files if there are multiple of them. With this information any data engineer can prepare a DAG that will run this project in production Airflow with requested schedule because almost all data science projects have common and simple code as a result of abstraction. By keeping project level domain knowledge in the notebook files and abstracting common tasks into Airflow operators, we have made scheduling a data science project task on Airflow a configuration process rather than a development effort.
Now we have talked about the way of working, let’s dive into details of system architecture and implementation.
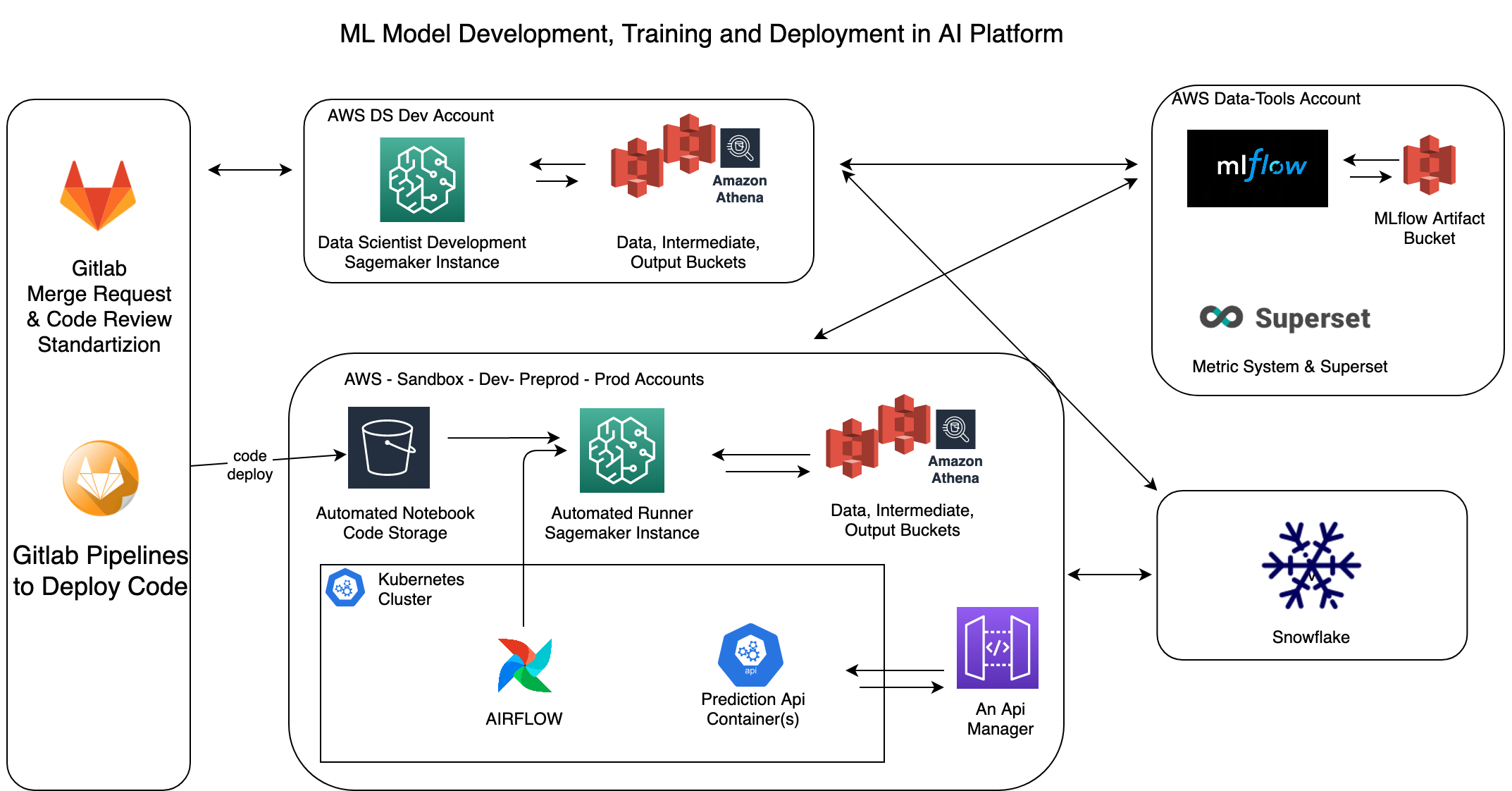
Above you can see AI Platform’s simplified overview diagram. Main components that we will discuss here are Airflow and Sagemaker but let me briefly explain some other tools as well.
We use mainly S3 buckets as input source to data science and each account has 3 main buckets on top of many other S3 buckets for other purposes.
- data, a read only bucket, contains parquet files in a folder structure that is similar to a Hive filesystem. We also provide AWS Athena on top of these files for ad-hoc queries.
- intermediate, read/write bucket, storage for temporary, short term files that are not important. Think like parquet or json outputs of a spark job that will be input for the next step in the pipeline.
- output, read/write bucket, storage for final products of applications. So prediction results, trained models or artifacts are stored here for the long term if necessary.
We also use MLflow in most of the projects, it is used for storing model artifacts and metrics across accounts. And Snowflake is the main data warehouse in the company, for some projects also used as input data source and for many projects Snowflake is the destination of output data of ML models. Loading data into Snowflake is done via Airflow.
If we go back to Airflow and Sagemaker orchestration for running notebook files, we can start by talking about how notebooks and other code in the project is deployed. As you can see in the diagram, each project’s code is stored in Gitlab as usual and the deployment task is actually doing one simple thing, copying the whole code in this project into a S3 bucket, which I call that bucket as automated-notebook. This bucket exists in each aws account with a suffix in the name, like other S3 buckets. So when you click deploy to preprod in gitlab pipeline, what it actually does is that copying project content into xx-automated-notebook-preprod bucket so that Sagemaker instance can access latest deployment version of the code in that branch when it is running the notebooks.
Ok, we have code in a place that is accessible to Sagemaker instance, now we need a trigger, which in our case Airflow. As I mentioned above, each project has a DAG and when the dag runs it executes almost standard tasks for every project, such as;
Create a Sagemaker instance
Based on DAG's requirements, we can run all notebooks in one sagemaker instance or we can run them in parallel using multiple sagemaker instances. Think like a scenario where you train a model for each country, so we can simply start 20 sagemaker instances for 20 countries at the same time, because there is no need to wait for each other training’s completion to start the next one. We usually do this and keep the total runtime of the dag same, even though the number of countries are increasing by the time. At some point you have to create AWS limit increase support tickets for each type of instance to increase your active Sagemaker instance count.
Get IP address of Sagemaker instance
Since we create a new instance each time, this also means that they got assigned new private ip addresses in the aws private subnet and we need this IP address to be able ssh into these instances. Although AWS is not returning this IP in boto3’s describe instance api response, since instances are created in our subnet, we are still able to find them in the elastic network interface (ENI) console. So with some boto3 api call trick, we are now able to get IP and pass it to the next task in Airflow via Xcom.
Run Notebooks via Papermill
To run papermill commands we will use Airflow SSHOperator with a couple of commands chained together.
- Checkout project code into newly created Sagemaker instance. This is an
aws s3 synccommand from automated-notebook bucket to local disk. - Activate your preferred conda environment that is used in this project.
- Install desired papermill version, since not every conda env has it by default.
- Install project dependencies via
pip install -r requirements.txtusing the file in the project. - Finally, with everything else is ready, we run the papermill command with notebook name, output destination, kernel and many other custom parameters that you want to pass to the notebook file.
Papermill is a great project and I like the 2 features of it most. Of course, the first one is injecting parameters. Papermill enables you to inject parameters into notebooks files if you just add a cell with parameters tag in ipynb file as described here. We are using this feature to pass a few standard parameters to notebooks such as aws environment name or id, the execution date from airflow dag in addition to project/model specific parameters.
Second feature of papermill is that you can save each notebook run file together with cell outputs and injected parameters and everything. Saved .ipynb file looks the same as if you would run it manually. This allows us two things, troubleshooting with detailed error logs and saving each scheduled run for audit or keeping history purposes. Basically each notebook file that ran in an automated way is kept on S3 as a separate file.
Delete Sagemaker instance
When all notebook files are executed via SSHOperator in Airflow dag, the next step is deleting this Sagemaker instance because we do not need it anymore. Next run of this dag and also other dags will create their own Sagemaker instance to run notebooks, this gives us a temporary and isolated runtime for each project as well as higher number of concurrent runners for projects. AWS Sagemaker management api limit is 4 requests per second, so you will need to handle that if you get api errors.
Delivering results
Since this flow is used for ML model training and also predictions/inference, most of the time dags will have a final step where we load output of notebooks to Snowflake or other destinations. These are json or parquet files based on project requirements and in our case interacting with Snowflake or other 3rd parties is much easier to do on the Airflow side than doing this in notebook files. Airflow has an operator or plugin for almost everything that we integrate with already, so we choose not to include this responsibility in data science project notebooks to keep it simple.
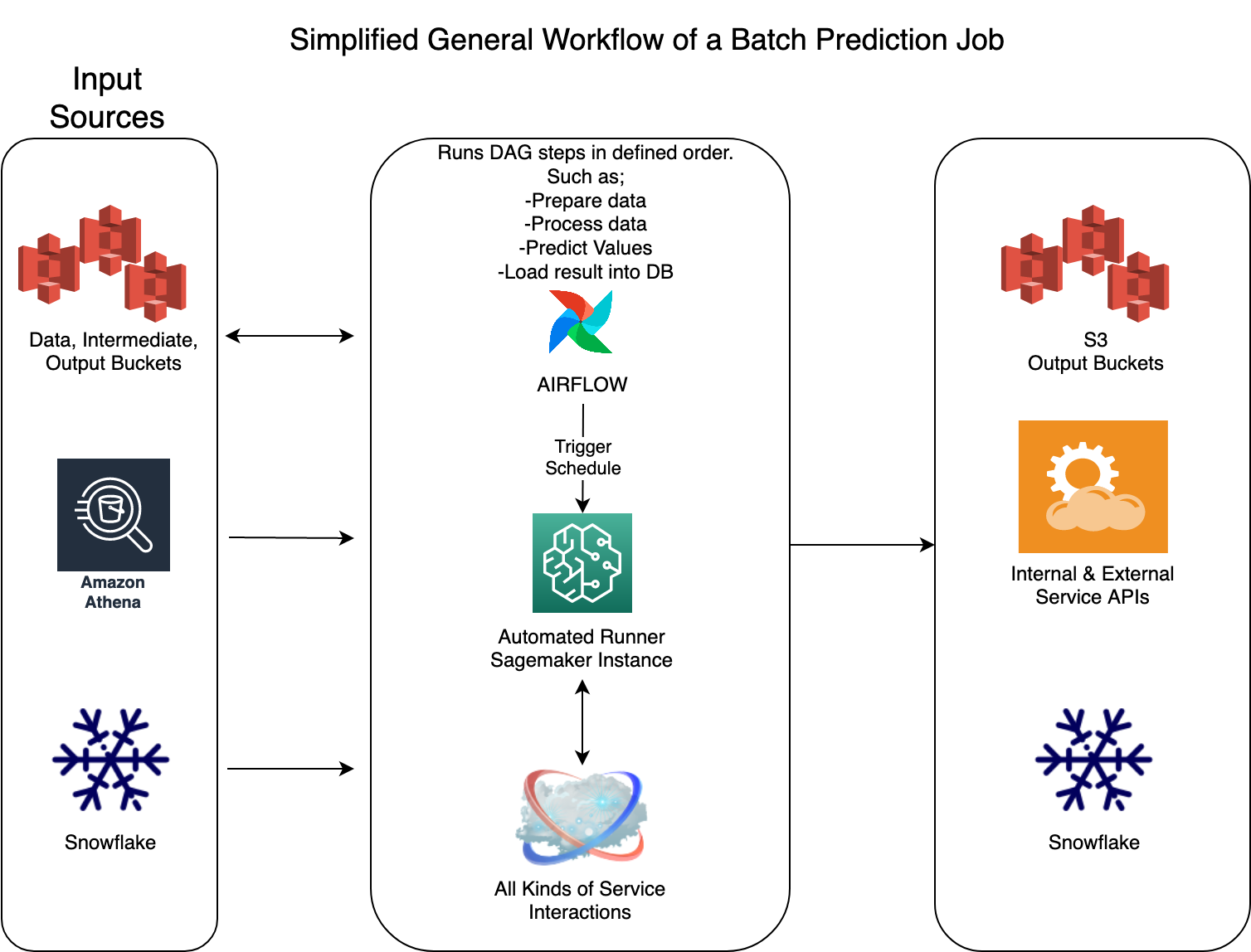
Above, I have added a simple flow for a typical Airflow DAG with Sagemaker notebook instance. Each notebook might be doing a very complex ML model training or prediction in it, but from Data Engineering perspective this is a simple application that is triggered via Airflow with parameters, interacting with variety of source data systems and producing an output which will be delivered to the destination system.
Benefits of this solution
- Data engineer does not have to know about the details of a data science project to automate it.
- Data scientist can develop and test a notebook without hard-coded parameters and will know that the same code is running on production, there is no copy pasting, no temporary line of codes etc. All code changes and history is on git repository.
- Application runtimes are same and isolated. Each sagemaker instance installs project requirements and runs only one project. There are no long living runtimes that can cause conflict with library versions or needs regular maintenance.
- You do not have to wait for another project to finish so the other project can start running.
- All Sagemaker instances are only used during application runtime, later terminated. So no cost for idle runners.
- Multiple software development environments can be used for testing or troubleshooting production issues since all instances are set up identical and have the same code from git repository.
- Any supported language or kernel can be used in a project and this is just a parameter in Airflow dag.
- Can keep a history of logs on Airflow and also in S3 from papermill output.
- There is no need for extensive docker experience or kubernetes cluster administrator.
- No need to deal with EMR cluster setup and related issues that come with it.
- You are limited with the largest Sagemaker instance type, currently 96 cpu and 768 GB of memory, but in our case this is enough and in return we do not have to maintain an EMR cluster or other runtimes to run data science workloads. And if needed there are alternative solutions to use while still using other benefits.
Links